The COVID-19 pandemic has deeply transformed our lives in the past few months. Fortunately, governments and researchers are still working hard to fight it.
In this post, I modestly bring my piece to the building and propose an AI-powered tool to help medical practitioners keep track of the latest research around COVID-19.
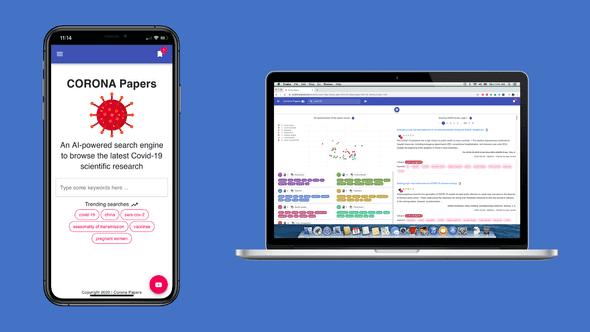
Without further ado, meet Corona Papers 🎉
In this post, I’ll:
- Go through the main functionalities of Corona Papers and emphasize what makes it different from other search engines
- Share the code of the data preprocessing and topic detection pipelines so that it can be applied in similar projects
What is Corona Papers?
Corona Papers is a search engine that indexes the latest research papers about COVID-19.
If you’ve just watched the video, the following sections will dive into more details. If you haven't watched it yet, all you need to know is here.
1 — A curated list of papers and rich metadata 📄
Corona Papers indexes the COVID-19 Open Research Dataset (CORD-19) provided by Kaggle. This dataset is a regularly updated resource of over 138,000 scholarly articles, including over 69,000 with full text, about COVID-19, SARS-CoV-2, and related coronaviruses.
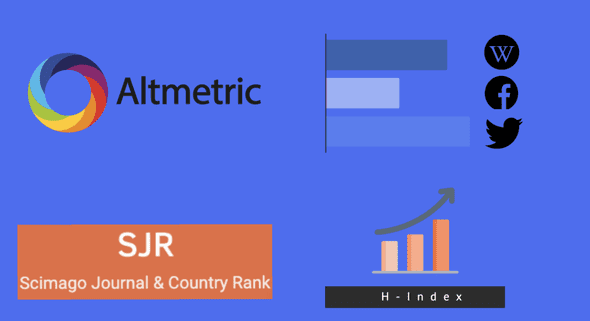
Corona Papers also integrates additional metadata from Altmetric and Scimgo Journal to account for the online presence and academic popularity of each article. More specifically, it fetches information such as the number of shares on Facebook walls, the number of posts on Wikipedia, the number of retweets, the H-Index of the publishing journal, etc.
The goal of integrating this metadata is to consider each paper’s impact and virality among the community, both the academic and non-academic ones.
2 — Automatic topic extraction using a language model 🤖
Corona Papers automatically tags each article with a relevant topic using a machine learning pipeline.
This is done using CovidBERT: a state-of-the-art language model fine-tuned on medical data. With the great power of the Hugging Face library, using this model is pretty easy.
Let’s break down the topic detection pipeline for more clarity:
- Given that abstracts represent the main content of each article, they’ll be used to discover the topics instead of the
full content. They are first embedded using CovidBERT. This produces vectors of 768 dimensions.
— Note that CovidBERT embeds each abstract as a whole so that the resulting vector encapsulates the semantics of the full document. - Principal Component Analysis (PCA) is performed on these vectors to reduce their dimension in order to remove redundancy and speed-up later computations. 250 components are retained to ensure 95% of the explained variance.
- KMeans clustering is applied on top of these PCA components in order to discover topics. After many iterations on the
number of clusters, 8 seemed to be the right choice.
— There are many ways to select the number of clusters. I personally looked at the silhouette plot of each cluster (figure below).
⚠️ An assumption has been made in this step: each article is assigned a unique topic, i.e. the dominant one. If you're looking at generating a mixture of topics per paper, the right way would be to use Latent Dirichlet Allocation. The downside of this approach, however, is that it doesn’t integrate COVIDBert embeddings. - After generating the clusters, I looked into each one of them to understand the underlying sub-topics. I first tried a
word-count and TF-IDF scoring to select the most important keywords per
cluster. But what worked best here was extracting those keywords by performing an LDA on the documents of each
cluster. This makes sense because each cluster is itself a collection of sub-topics.
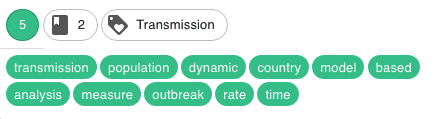
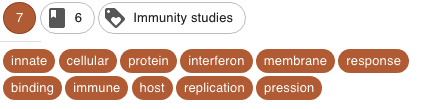
Different coherent clusters were discovered. Here are some examples, with the corresponding keywords (the cluster names have been manually attributed on the basis of the keywords)

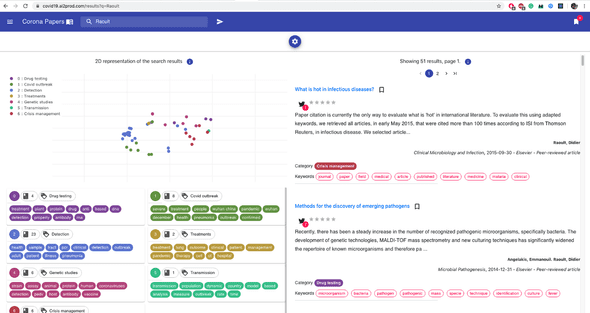
- I decided, finally, and for fun mainly, to represent the articles in an interactive 2D map to provide a visual interpretation of the clusters and their separability. To do this, I applied a tSNE dimensionality reduction on the PCA components. Nothing fancy.
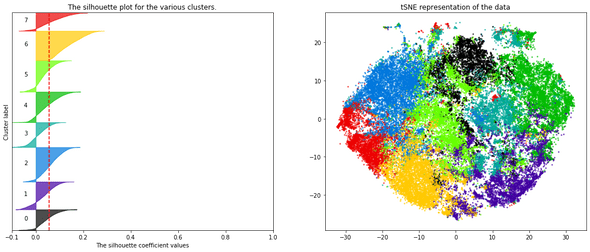
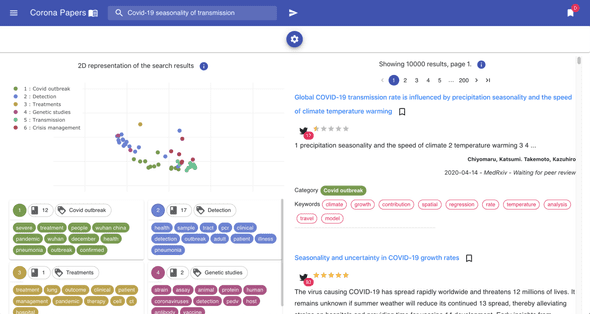
To bring more interactivity to the search experience, I decided to embed the tSNE visualization into the search results (this is available on Desktop view only).
On each result page, the points on the plot (on the left) represent the same search results (on the right): this gives an idea on how results relate to each other in a semantic space.
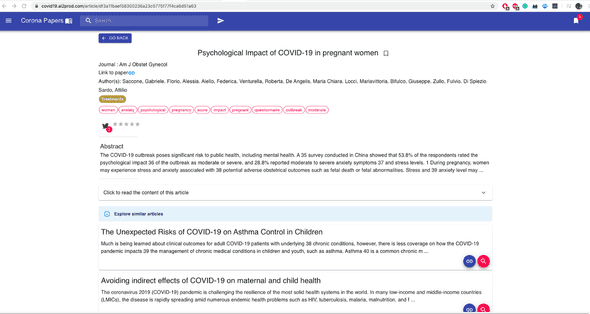
3 — Recommendation of similar papers
Once you click on a given paper, Corona Papers with show you detailed information about it such as the title, the abstract, the full content, the URL to the original document, etc.
Besides, it proposes a selection of similar articles that the user can read and bookmark.
These articles are based on a similarity measure computed on CovidBert embeddings. Here are two examples:

4 — A stack of modern web technologies 📲
Corona Papers is built using modern web technologies

Back-end
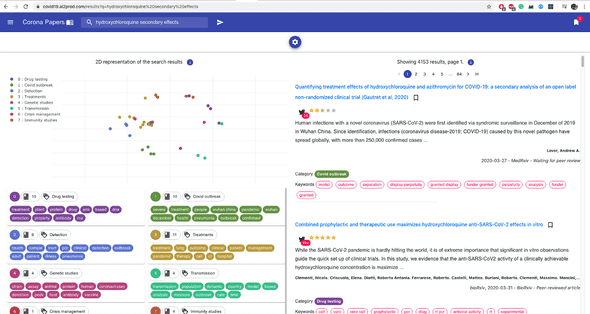
At its core, it uses Elasticsearch to perform full-text queries, complex aggregation and sorting. When you type in a list of keywords, for example, Elasticsearch matches them first with the titles, abstract, and eventually the author names.
Here’s an example of a query:
And here’s a second one that matches an author’s name:
The next important component of the backend is a Flask API: it makes the interface between ElasticSearch and the front-end.
Front-end
The front-end interface is built using Material-UI, a great React UI library with a variety of well-designed and robust components.
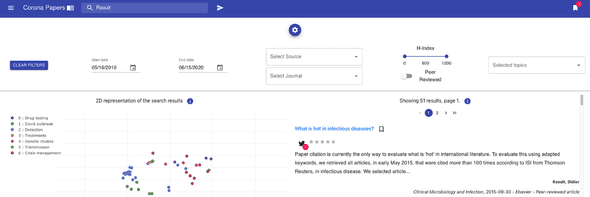
It has been used to design the different pages, and more specifically the search page with its collapsable panel of search filters :
- publication date
- publishing company (i.e. the source)
- journal name
- peer-reviewed articles
- h-index of the journal
- the topics
Because accessibility matters, I aimed at making Corona Papers a responsive tool that researchers can use on different devices. Using Material-UI helped us design a clean and simple interface.

Cloud and DevOps
I deployed Corona Papers on AWS using docker-compose.
How to use CovidBERT in practice
Using the sentence_transformers package to load and generate embedding from CovidBERT is as easy writing these few lines
import os
import numpy as np
from sentence_transformers import SentenceTransformer res
# initialize the model
model = SentenceTransformer("./src/models/covidbert/")
# generate embeddings
excerpt_embeddings = model.encode(excerpts, show_progress_bar=True, batch_size=32)
excerpt_embeddings = np.array(excerpt_embeddings)
np.save(os.path.join(export_path, "embeddings_excerpts.npy"), excerpt_embeddings)If you’re interested in the data processing and topic extraction pipelines, you can look at the code in my Github repository.
You’ll find two notebooks:
1-data-consolidation.ipynb:
- consolidates the CORD database with external metadata from Altmetric, Scimago Journal, and CrossRef
- generates CovidBERT embeddings from the titles and excerpts
2-topic-mining.ipynb:
- generates topics using CovidBERT embeddings
- select relevant keywords for each cluster
What key lessons can be learned from this project?
Building Corona Papers has been a fun journey. It was an opportunity to mix up NLP, search technologies, and web design. This was also a playground for a lot of experiments.
Here are some technical and non-technical notes I first kept to myself but am now sharing with you:
- Don’t underestimate the power of Elasticsearch. This tool offers great customizable search capabilities. Mastering it
requires a great deal of effort but it’s a highly valuable skill.
Visit the official website to learn more. - Using language models such as CovidBERT provides efficient representations for text similarity tasks.
If you’re working on a text similarity task, look for a language model that is pretrained on a corpus that resembles yours. Otherwise, train your own language model.
There are lots of available models here. - Docker is the go-to solution for deployment. Pretty neat, clean, and efficient to orchestrate the multiple services of
your app.
Learn more about Docker here. - Composing a UI in React is really fun and not particularly difficult, especially when you play around with libraries
such as Material-UI.
The key is to first start by sketching your app, then design individual components separately, and finally assemble the whole thing.
This took me a while to grasp because I was new to React, but here are some tutorials I used: — React official website
— Material UI official website where you can find a bunch of components
— I also recommend this channel. It’s awesome, fun, and quickly gets you to start with React fundamentals. - Text clustering is not a fully automatic process. You’ll have to fine-tune the number of clusters almost manually to find the right value. This requires monitoring some metrics and qualitatively evaluating the results.
Of course, there are things I wish I had time to try like setting up CI-CD workflow with Github actions and building unit tests. If you have experience with those tools, I’d really appreciate your feedback.
Spread the word! Share Corona Papers with your community
If you made it this far, I’d really want to thank you for reading!
If you find Corona Papers useful, please share this link: https//covid19.ai2prod.com with your community.
If you have a feature request for improvement or if you want to report a bug, don’t hesitate to contact me.
I’m looking forward to hearing from you! Best,